Task 1
You are required to perform the process of normalising the data shown in Appendix A and B to first (1NF), second (2NF) and third (3NF). Ensure you clearly outline the process you went
through to create the database in 3NF and identity the appropriate primary and foreign keys.
Task 2
You are required to create an Oracle database which will support the queries listed below for the database your have designed in Task 1. You should select appropriate data types for
each of the attributes in the tables. The database should be populated with the sample data provided in Appendix B. If you wish you can add additional data yourself.
Clearly you will have to deduce the data from the table (Appendix B) that should be entered in the individual tables you created in Task 1 and insert additional information when required.
Task 3
In no more than 750 words, create a report that identifies how the use of cloud computing and Big Data analysis could aid the Hippocratic medical centre and similar health related
institutions. You should identify suitable applications, and consider the benefits that could be achieved through the introduction of cloud computing and Big Data analysis, and any
restriction that need to be overcome to achieve their introduction. Your report should be referenced using the appropriate CU Harvard approach.
Database design is the process of producing a detail structural model of a database. A fully designed database comprises of the detailed structures of all the entities with their attributes, definitions, and the relationships among them.
The overall database system consists of the designs and definitions of many different parts of the database. Most commonly, the logical design of the base data structures show what data to store and where to store which data.
The given table in Appendix B is not in a normalised form, which means there might be updation, deletion, insertion anomalies or redundancies of data existing in that table. In order to get rid of these anomalies and redundant data we need to normalise this database. In order to avoid relation, analyst divides this relation into smaller relations.
The table can be divided into following relations. Given below are the table definitions, their field names, data types, field sizes and primary key and foreign key specifications:
|
Field |
Data type |
Key |
|
P_id |
Number(5) |
Primary key |
|
Fname |
Varchar2(20) |
|
|
Lname |
Varchar2(20) |
|
|
Ward |
Number(4) |
|
|
Doc_id |
Number(5) |
Foreign key |
|
Phar_id |
Number(5) |
Foreign key |
|
Field |
Data type |
Key |
|
Doc_id |
Number(5) |
Primary key |
|
Name |
Varchar2(30) |
|
Field |
Data type |
Key |
|
Phar_id |
Number(5) |
Primary key |
|
Name |
Varchar2(30) |
|
Field |
Data type |
Key |
|
Med_id |
Number(5) |
Primary key |
|
Name |
Varchar2(30) |
|
|
Dosage |
Varchar2(20) |
|
|
Side_effect |
Varchar2(50) |
|
Field |
Data type |
Key |
|
Start |
Date |
|
|
End |
Date |
|
|
Med_id |
Number(5) |
Foreign key |
|
P_id |
Number(5) |
Foreign key |
|
Doc_id |
Number(5) |
Foreign key |
[Note: - The tables are defined using the Oracle 11g built-in data types and are Oracle- supported data types. It will change in MySQL or other database languages.]
There are several steps involved in the designing of the database. Namely:
- Gathering of requirements and analysis
- Conceptual database design (E-R Diagram)
- Logical database design (table definitions, normalization, functional dependencies etc.)
- Physical Database design (Clustering, indexing etc.)
- This database design is normalized up to 3NF.
- By seeing the Relational Model (Table definition) we can easily develop the E-R Model and vice versa is true, which basically means steps 2 and 3 are interchangeable.
- Entity integrity and Referential integrity rule is set properly.
- Domain integrity can be set easily.
|
Normalisation is the process of dividing or breaking one or more relations to a group of smaller relations in order to get rid of data anomalies, data redundancies and to ensure data integrity. Data redundancy means existence of duplicate, or similar data in several places in the database. Mainly redundancies exist when there is unnecessary duplicity of data in different relations inside the database. Data integrity suggests that all of the data in the database strictly follow all the integrity constraints and maintain data consistency. The data in the database can be in one or more ‘normal forms’. Strictly, there are 3 basic, most common and most important normal forms. These follow a strict order: · 1st normal form · 2nd normal form · 3rd normal form |
First Normal Form suggests that data must not be repeated in any two or more rows of a table or relation. There should not be any repeating information and each set of domain of column must contain a unique atomic value.
If we break the table given in appendix B in the following way, there are three different tables representing every patient’s details, the general medicine details and the medical histories of each patient respectively.
(i) Patient {p_id, fname, lname, ward, doc_id, phar_id}
Task 2: Creating an Oracle database
(i) Medication {med_id, name, dosage, side_effect}
(iii) Medical_history{p_id, med_id, doc_id, start, end}
The primary keys for the tables are respectively p_id, med_id, and for the 3rd table the concatenated key {p_id, med_id}.
In the above three tables, there are no repeating elements in any of the fields.
Hence it is normalised to 1NF.
Second Normal Form suggests that partial dependency on primary key should not exist for any column or attribute. If a relation has concatenated primary key, every non-prime attribute in the table must be dependent upon the entire concatenated key and not on any part of it.
The first two tables (patient table and medication table) have only one field as their primary key. Thus they have no cases of partial dependency.
However for the 3rd table (medicine history table) there is a concatenated key {p_id, med_id}.
Here the non-key attributes {start} and {end} are fully dependent on the whole of the concatenated key and not part of it. Thus there are no partial dependencies.
Hence it is normalised up to 2NF.
Third Normal form suggests that every non-prime attribute of a relation must depend only on the primary key of that relation, or in other way, there should be no existence of any non-prime attribute in the table which is determined by another non-prime attribute of that table.
In table no. (i): patient{p_id, fname, lname, ward, doc_id, pha_id} every non-prime attribute depends on {p_id} which is the primary key.
In table no (ii): medication {med_id, name, dosage, side_effect} every non-prime attribute depends on {med_id} which is the praimary key.
In table no. (iii): medical_history {p_id, med_id, doc_id, start, end} every non-prime attribute depends on {p_id, med_id} which is the concatenated primary key.
Hence it is normalised up to 3NF.
Thus we can say that all the relations in this database follow 1NF, 2NF and 3NF.
The ER-Diagram shows the specified entities and relationship among those entities. The attributes belonging to each entity are shown, including the identifier attributes (primary keys). The relationship cardinalities are also shown in the diagram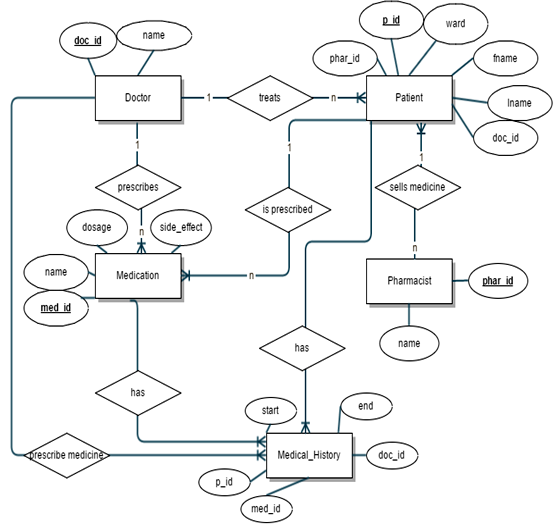
There are mainly four entities in the database design, namely,
Patient, doctor, pharmacist, medication and medical_history. These entities are represented as a rectangular box in the ER diagram. Each entity has a set of attributes. The attributes are the fields in the specific tables. Attributes in the ER-diagram are represented inside an oval shape. Each of the relations in the database has a particular attribute or field which helps to uniquely identify each row or record in that table. This attribute is called the identifier attribute or primary key of that table or relation.
The attributes which matches with another attributes in a different relation are the foreign keys. A foreign key in one table uniquely identifies a record in another table. In other words, the foreign key is that column in the child table that refers to the primary key in the parent table.
In table no. (i), patient table: {p_id} is identified as the primary key of the table as it uniquely identifies each patient.
The fields: doc_id and phar_id are foreign keys from tables doctor and pharmacist respectively.
In table no. (ii), doctor table: {doc_id} is identified as the primary key as it uniquely determines each doctor record.
Task 3: Cloud computing and Big Data analysis in healthcare
In table no. (iii), pharmacist table: {phar_id} is identified as the primary key as it uniquely determines each pharmacist record.
In table no. (iv), medication table: {med_id} is set as the primary key as it uniquely identifies each medicine detail record.
After these tables are created and populated with the sample data that has been provided in Appendix B, the tables will look like this when SQL SELECT statement is run on them. The view or output is presented below for quick reference.
|
P_id |
Fname |
Lname |
Ward |
Doc_id |
Phar_id |
|
0501 |
Shila |
Smith |
22 |
D123 |
P001 |
|
0102 |
Mark |
Shaw |
01 |
D123 |
P003 |
|
0001 |
Graeme |
Smith |
23 |
D002 |
P001 |
|
0002 |
Diane |
Price |
22 |
D003 |
P002 |
|
0051 |
Julie |
Dixon |
33 |
D002 |
D003 |
|
0031 |
Cornelius |
Bower |
21 |
D022 |
P004 |
|
0013 |
Ethel |
Weber |
11 |
D012 |
P033 |
|
0012 |
Caroline |
Garen |
11 |
D013 |
P001 |
|
0014 |
John |
Malone |
10 |
D032 |
P033 |
|
Doc_id |
Name |
|
D123 |
Stewart |
|
D002 |
Elshaw |
|
D003 |
Smith |
|
D022 |
Arevian |
|
D012 |
Jabok |
|
D013 |
Yi |
|
D032 |
Murray |
|
Phar_id |
Name |
|
P001 |
Hamshaw |
|
P003 |
Sallis |
|
P002 |
Jennis |
|
P004 |
Moore |
|
P033 |
Holt |
|
Med_id |
Name |
Dosage |
Side_effect |
|
1001 |
Citalopram |
20mg |
Memory loss |
|
1004 |
Jetrea |
0.125mg |
Decreased vision |
|
1010 |
Codeine |
10mg |
Confusion |
|
1009 |
Geodon |
20mg |
Cough |
|
1005 |
Kelfex |
4g |
Diarreah |
|
1011 |
Pristiq |
50mg |
Cold chills |
|
1007 |
Zofran |
24mg |
Confusion |
|
1012 |
Lisinopril |
10mg |
Blurred vision |
|
1013 |
Asprin |
75mg |
Bleeding |
|
1020 |
Prozac |
20mg |
None |
|
1111 |
Jelltrears |
5mg |
Blurred vision |
|
Med_id |
P_id |
Doc_id |
Start |
End |
|
1001 |
0501 |
D123 |
22/02/2014 |
25/04/2014 |
|
1004 |
0501 |
D123 |
22/05/2014 |
01/06/2014 |
|
1010 |
0102 |
D123 |
03/09/2013 |
06/08/2014 |
|
1009 |
0102 |
D123 |
04/09/2013 |
06/09/2013 |
|
1001 |
0001 |
D002 |
03/05/2012 |
03/10/2013 |
|
1005 |
0001 |
D002 |
06/06/2008 |
09/04/2010 |
|
1011 |
0001 |
D002 |
07/10/2011 |
22/12/2011 |
|
1004 |
0001 |
D002 |
22/06/2014 |
11/09/2014 |
|
1007 |
0002 |
D003 |
11/04/2005 |
12/08/2007 |
|
1010 |
0002 |
D003 |
03/07/2013 |
06/10/2013 |
|
1012 |
0051 |
D002 |
17/06/2011 |
17/05/2012 |
|
1004 |
0051 |
D002 |
22/05/2014 |
01/06/2014 |
|
1013 |
0033 |
D022 |
12/04/2012 |
23/07/2012 |
|
1020 |
0033 |
D022 |
01/04/1970 |
12/02/2010 |
|
1013 |
0013 |
D012 |
07/04/2014 |
11/04/2014 |
|
1020 |
0013 |
D012 |
03/05/2012 |
03/10/2013 |
|
1004 |
0012 |
D013 |
22/06/2014 |
11/09/2014 |
|
1111 |
0012 |
D013 |
21/05/2012 |
12/06/2012 |
|
1020 |
0014 |
D032 |
01/04/01970 |
12/02/2010 |
|
1010 |
0014 |
D032 |
03/08/2014 |
06/10/2014 |
FROM patient p INNER JOIN doctor d
ON p.doc_id=d.doc_id JOIN pharmacist ph
ON p.phar_id=ph.phar_id;
Output:
|
Fname |
Lname |
ward |
Doc_name |
Phar_name |
|
Caroline |
Garen |
22 |
Stewart |
Hamshaw |
|
Cornelius |
Bower |
01 |
Elshaw |
Sallis |
|
Diane |
Price |
23 |
Smith |
Jennis |
|
Ethel |
Weber |
22 |
Arevian |
Moore |
|
Graeme |
Smith |
33 |
Jabok |
Holt |
|
John |
Malone |
21 |
Yi |
Jennis |
|
Julie |
Dixon |
11 |
Murray |
Moore |
|
Mark |
Shaw |
11 |
Jabok |
Jennis |
SELECT p.f_name, p.l_name, p.ward, mh.med_id, mh.start. mh.end
FROM patient p INNER JOIN doctor d
ON p.doc_id=d.doc_id JOIN medicine_history mh
ON p.patient_id=mh.patient_id
WHERE d.name=”Elshaw”;
Output:
|
Fname |
Lname |
Ward |
Med_name |
|
Graeme |
Smith |
23 |
Cilatopram |
|
Graeme |
Smith |
23 |
Kelfex |
|
Julie |
Dixon |
33 |
Lisinopril |
|
Julie |
Dixon |
33 |
Jetrea |
SELECT p.f_name, p.l_name, ph.name
FROM patient p INNER JOIN pharmacist ph
ON p.phar_id=ph.phar_id JOIN med_history mh
ON p.patient_id=mh.patient_id
WHERE mh.start<’01/01/2010’;
|
Fname |
Name |
Start |
|
Graeme |
Kelfex |
06/06/2008 |
|
Diane |
zofran |
11/04/2005 |
|
Caroline |
Prozac |
1/4/1970 |
|
John |
Prozac |
1/4/1970 |
SELECT d.name, m.name
FROM doctor d INNER JOIN medication m
ON d.doc_id=m.doc_id
WHERE m.name=’Aspirin’ OR m.name=’Codeine’;
|
Name |
Name |
|
Stewart |
Codeine |
|
Smith |
Codeine |
|
Arevian |
Asprin |
|
Murray |
Codeine |
SELECT count(*) FROM patient p INNER JOIN medical_history mh
ON p.p_id=mh.p_id WHERE mh.start>=’01/01/2014’ AND mh.start<=’31/12/2014’;
SELECT d.name, p.fname, p.lname
FROM patient p INNER JOIN doctor d
ON p.doc_id=d.doc_id JOIN medical_history mh
ON mh.p_id=p.p_id
WHERE mh.start>=’01/01/2014’ AND mh.start<=’31/12/2014’;
|
Fname |
Name |
|
Shila |
Stewart |
|
Gareme |
Smith |
|
caroline |
Yi |
Cloud computing is the idea of shifting computer services such as computation and information storage to one or more redundant offsite locations which might be available on the Internet (Chard, Tuecke and Foster, 2014). This helps us to use and operate any application software through internet-enabled devices.
Cloud computing is a type of computing which basically drives the sharing of large amount of data and information through shared digital resources rather than through individual local servers or personal computers (Collins, 2014). To some extent cloud computing is similar to grid computing. They are similar in the sense that all unused processing cycles of all systems that are connected via a network and can be utilized to solve problems which can be too intensive for small local servers or stand-alone machines (Nepal and Pandey, 2015).
Cloud computing can be of great use for the Hippocratic Medical Centre because of the way it works (Plunkett, 2014). Users can be benefited by the use of clouds in following ways:
Reduced costs: The price of moving applications in the cloud is less than that of any on-site machine due to the minimum hardware costs because then the use of physical resources will be more effective (Sill, 2014).
This can be of serious help for this institute.
Common access: cloud computing makes it possible for the remotely located users or customers, who do not have that particular data or application software installed on their machine, to access those sites or applications via the internet (Yeo et al. 2012).
Up-to-date data: it becomes easier for the cloud provider to upgrade the software based on the feedback from customers who used the previous releases (Tari, 2014).
Ease of choosing application:By introducing clouds, the organisation can facilitate the cloud users and gives them the flexibility to experiment and choose the best option suited for them (Sasikala, 2013). Cloud computing also enables the organisation to use access and pay for only the products that they use and that too with a faster implementation time (Ranjan, 2014).
Can be greener and economical: In the cloud the average amount of energy which is needed for a singular computational action to be performed is far less than that of the average amount of energy for an on-site hosting (Nepal, Ranjan and Choo, 2015). This happens because different institutions and their branches can share the resources safely and securely, thus allowing the use of shared resources in a more efficient way.
Flexible: cloud computing facilitates cloud users to switch and swap applications easily and rapidly (Khurana, 2014). Thus it is easy for them to select the one that suits their needs the most.
Thus, if Hippocratic Medical Institution adopts this technology into their system their system would be more feasible, easy to operate and will get all the benefits of cloud.
Today, through the help of Big Data, large institutions and organisations are accessing data more than ever before. Previously, the data that were considered ‘dead’ or ‘of no value’, because they were unstructured or old, have now been collected, analysed and reused for the benefit of organisations (Huang and Nicol, 2013). Organisations have now been blessed with the opportunity of discovering correlations between data and matching patterns that were previously hidden (Buyya, 2013). Implementation of Big Data Analytics tools gives the organisations more power now as they can access more accurate data and information (Franks, 2012). It obviously helps and influences their business.
Big Data can provide benefits to an institution such as Hippocratic Medical Centre, in following ways:
Ensure there are more than enough data to make important decisions about business and those data are accurate and up-to-date (Choo, 2014). Big Data can play a major part in making this job far easier than the conventional manual way.
The amount of availability of data can be increasing using Big Data. It will improve marketing strategies and target more customers, thus increasing the customer base (Chen, Bhargava and Zhongchuan, 2014).
Ultimately, big data can be beneficial to an organisation as it is possible to increase revenue, reduce cost, attracts customers (Chard, Tuecke and Foster, 2014).
Organisations business decisions are improved making it possible to attract the large number of customers, cutting costs and developing smart marketing strategies (Catlett, 2013).
Thus, Big Data can largely contribute to these types of Institutions and ultimately increase their business prospects, provide a solid base for structuring the large amount records in the database, increase the availability of data and records.
Reference List:-
Bhargava, B., Khalil, I. and Sandhu, R. (2014). Securing Big Data Applications in the Cloud [Guest editors' introduction]. IEEE Cloud Computing, 1(3), pp.24-26.
Buyya, R. (2013). Introduction to the IEEE Transactions on Cloud Computing. IEEE Transactions on Cloud Computing, 1(1), pp.3-21.
Catlett, C. (2013). Cloud computing and big data. Amsterdam: IOS Press.
Chard, K., Tuecke, S. and Foster, I. (2014). Efficient and Secure Transfer, Synchronization, and Sharing of Big Data. IEEE Cloud Computing, 1(3), pp.46-55.
Chen, H., Bhargava, B. and Zhongchuan, F. (2014). Multilabels-Based Scalable Access Control for Big Data Applications. IEEE Cloud Computing, 1(3), pp.65-71.
Choo, K. (2014). Mobile Cloud Storage Users. IEEE Cloud Computing, 1(3), pp.20-23.
Collins, E. (2014). Big Data in the Public Cloud. IEEE Cloud Computing, 1(2), pp.13-15.
Franks, B. (2012). Taming the big data tidal wave. Hoboken, New Jersey: John Wiley & Sons, Inc.
Huang, J. and Nicol, D. (2013). Trust mechanisms for cloud computing. J Cloud Comput Adv Syst Appl, 2(1), p.9.
Jamshidi, P., Ahmad, A. and Pahl, C. (2013). Cloud Migration Research: A Systematic Review.IEEE Transactions on Cloud Computing, 1(2), pp.142-157.
Khurana, A. (2014). Bringing Big Data Systems to the Cloud. IEEE Cloud Computing, 1(3), pp.72-75.
Khurana, A. (2014). Bringing Big Data Systems to the Cloud. IEEE Cloud Computing, 1(3), pp.72-75.
Nepal, S. and Pandey, S. (2015). Guest Editorial: Cloud Computing and Scientific Applications (CCSA)--Big Data Analysis in the Cloud. The Computer Journal.
Nepal, S., Ranjan, R. and Choo, K. (2015). Trustworthy Processing of Healthcare Big Data in Hybrid Clouds. IEEE Cloud Computing, 2(2), pp.78-84.
Plunkett, J. (2014). Plunkett's InfoTech Industry Almanac 2014. Houston: Plunkett Research, Ltd.
Ranjan, R. (2014). Modeling and Simulation in Performance Optimization of Big Data Processing Frameworks. IEEE Cloud Computing, 1(4), pp.14-19.
Sasikala, P. (2013). Energy efficiency in cloud computing: way towards green computing. IJCC, 2(4), p.305.
Sill, A. (2014). Cloud Standards and the Spectrum of Development. IEEE Cloud Computing, 1(3), pp.15-19.
Tari, Z. (2014). Security and Privacy in Cloud Computing. IEEE Cloud Computing, 1(1), pp.54-57.
Yeo, S., Pan, Y., Lee, Y. and Chang, H. (2012). Computer science and its applications. Dordrecht: Springer.
To export a reference to this article please select a referencing stye below:
My Assignment Help. (2017). Database Design And Normalization For Hippocratic Medical Centre. Retrieved from https://myassignmenthelp.com/free-samples/data-normalising-process.
"Database Design And Normalization For Hippocratic Medical Centre." My Assignment Help, 2017, https://myassignmenthelp.com/free-samples/data-normalising-process.
My Assignment Help (2017) Database Design And Normalization For Hippocratic Medical Centre [Online]. Available from: https://myassignmenthelp.com/free-samples/data-normalising-process
[Accessed 01 June 2025].
My Assignment Help. 'Database Design And Normalization For Hippocratic Medical Centre' (My Assignment Help, 2017) <https://myassignmenthelp.com/free-samples/data-normalising-process> accessed 01 June 2025.
My Assignment Help. Database Design And Normalization For Hippocratic Medical Centre [Internet]. My Assignment Help. 2017 [cited 01 June 2025]. Available from: https://myassignmenthelp.com/free-samples/data-normalising-process.
